Detection of quality deviations in paracetamol products
Fusion of Mass Spectrometry and Raman Spectroscopy
Data
Ricardo Cunha
cunha@iuta.de01 August, 2024
Source:vignettes/articles/demo_paracetamol.Rmd
demo_paracetamol.RmdPre-processing
MS
| analysis | replicate | blank | description |
|---|---|---|---|
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-39-1_091 | ass_1 | blank | Aspirin standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-39-1_092 | ass_1 | blank | Aspirin standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-39-1_093 | ass_1 | blank | Aspirin standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-39-2_094 | ass_2 | blank | Aspirin standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-39-2_095 | ass_2 | blank | Aspirin standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-39-2_096 | ass_2 | blank | Aspirin standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-39-3_097 | ass_3 | blank | Aspirin standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-39-3_098 | ass_3 | blank | Aspirin standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-39-3_099 | ass_3 | blank | Aspirin standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_Blank_100 | blank | blank | Reference background |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_Blank_101 | blank | blank | Reference background |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_Blank_111 | blank | blank | Reference background |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_Blank_112 | blank | blank | Reference background |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_Blank_009 | blank_e | blank_e | Reference background |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_Blank_010 | blank_e | blank_e | Reference background |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_Blank_020 | blank_e | blank_e | Reference background |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_Blank_021 | blank_e | blank_e | Reference background |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_Blank_031 | blank_e | blank_e | Reference background |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_Blank_032 | blank_e | blank_e | Reference background |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_Blank_042 | blank_e | blank_e | Reference background |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_Blank_043 | blank_e | blank_e | Reference background |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_Blank_053 | blank_e | blank_e | Reference background |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_Blank_054 | blank_e | blank_e | Reference background |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-40-1_102 | caff_1 | blank | Caffeine standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-40-1_103 | caff_1 | blank | Caffeine standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-40-1_104 | caff_1 | blank | Caffeine standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-40-2_105 | caff_2 | blank | Caffeine standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-40-2_106 | caff_2 | blank | Caffeine standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-40-2_107 | caff_2 | blank | Caffeine standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-40-3_108 | caff_3 | blank | Caffeine standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-40-3_109 | caff_3 | blank | Caffeine standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-40-3_110 | caff_3 | blank | Caffeine standard |
| 20240415-31-1_003 | fiz_a1 | blank | Paracetamol product |
| 20240415-31-1_004 | fiz_a1 | blank | Paracetamol product |
| 20240415-31-1_005 | fiz_a1 | blank | Paracetamol product |
| 20240415-31-2_006 | fiz_a2 | blank | Paracetamol product |
| 20240415-31-2_007 | fiz_a2 | blank | Paracetamol product |
| 20240415-31-2_008 | fiz_a2 | blank | Paracetamol product |
| 20240415-31-3_009 | fiz_a3 | blank | Paracetamol product |
| 20240415-31-3_010 | fiz_a3 | blank | Paracetamol product |
| 20240415-31-3_011 | fiz_a3 | blank | Paracetamol product |
| 20240415-37-1_069 | fiz_acid1 | blank | Paracetamol with acetic acid |
| 20240415-37-1_070 | fiz_acid1 | blank | Paracetamol with acetic acid |
| 20240415-37-1_071 | fiz_acid1 | blank | Paracetamol with acetic acid |
| 20240415-37-2_072 | fiz_acid2 | blank | Paracetamol with acetic acid |
| 20240415-37-2_073 | fiz_acid2 | blank | Paracetamol with acetic acid |
| 20240415-37-2_074 | fiz_acid2 | blank | Paracetamol with acetic acid |
| 20240415-37-3_075 | fiz_acid3 | blank | Paracetamol with acetic acid |
| 20240415-37-3_076 | fiz_acid3 | blank | Paracetamol with acetic acid |
| 20240415-37-3_077 | fiz_acid3 | blank | Paracetamol with acetic acid |
| 20240415-35-1_047 | fiz_ass1 | blank | Paracetamol with aspirin |
| 20240415-35-1_048 | fiz_ass1 | blank | Paracetamol with aspirin |
| 20240415-35-1_049 | fiz_ass1 | blank | Paracetamol with aspirin |
| 20240415-35-2_050 | fiz_ass2 | blank | Paracetamol with aspirin |
| 20240415-35-2_051 | fiz_ass2 | blank | Paracetamol with aspirin |
| 20240415-35-2_052 | fiz_ass2 | blank | Paracetamol with aspirin |
| 20240415-35-3_053 | fiz_ass3 | blank | Paracetamol with aspirin |
| 20240415-35-3_054 | fiz_ass3 | blank | Paracetamol with aspirin |
| 20240415-35-3_055 | fiz_ass3 | blank | Paracetamol with aspirin |
| 20240415-32-1_014 | fiz_b1 | blank | Paracetamol product |
| 20240415-32-1_015 | fiz_b1 | blank | Paracetamol product |
| 20240415-32-1_016 | fiz_b1 | blank | Paracetamol product |
| 20240415-32-2_017 | fiz_b2 | blank | Paracetamol product |
| 20240415-32-2_018 | fiz_b2 | blank | Paracetamol product |
| 20240415-32-2_019 | fiz_b2 | blank | Paracetamol product |
| 20240415-32-3_020 | fiz_b3 | blank | Paracetamol product |
| 20240415-32-3_021 | fiz_b3 | blank | Paracetamol product |
| 20240415-32-3_022 | fiz_b3 | blank | Paracetamol product |
| 20240415-33-1_025 | fiz_c1 | blank | Paracetamol product |
| 20240415-33-1_026 | fiz_c1 | blank | Paracetamol product |
| 20240415-33-1_027 | fiz_c1 | blank | Paracetamol product |
| 20240415-33-2_028 | fiz_c2 | blank | Paracetamol product |
| 20240415-33-2_029 | fiz_c2 | blank | Paracetamol product |
| 20240415-33-2_030 | fiz_c2 | blank | Paracetamol product |
| 20240415-33-3_031 | fiz_c3 | blank | Paracetamol product |
| 20240415-33-3_032 | fiz_c3 | blank | Paracetamol product |
| 20240415-33-3_033 | fiz_c3 | blank | Paracetamol product |
| 20240415-34-1_036 | fiz_caff1 | blank | Paracetamol with caffeine |
| 20240415-34-1_037 | fiz_caff1 | blank | Paracetamol with caffeine |
| 20240415-34-1_038 | fiz_caff1 | blank | Paracetamol with caffeine |
| 20240415-34-2_039 | fiz_caff2 | blank | Paracetamol with caffeine |
| 20240415-34-2_040 | fiz_caff2 | blank | Paracetamol with caffeine |
| 20240415-34-2_041 | fiz_caff2 | blank | Paracetamol with caffeine |
| 20240415-34-3_042 | fiz_caff3 | blank | Paracetamol with caffeine |
| 20240415-34-3_043 | fiz_caff3 | blank | Paracetamol with caffeine |
| 20240415-34-3_044 | fiz_caff3 | blank | Paracetamol with caffeine |
| 20240415-36-1_058 | fiz_d1 | blank | Paracetamol product |
| 20240415-36-1_059 | fiz_d1 | blank | Paracetamol product |
| 20240415-36-1_060 | fiz_d1 | blank | Paracetamol product |
| 20240415-36-2_061 | fiz_d2 | blank | Paracetamol product |
| 20240415-36-2_062 | fiz_d2 | blank | Paracetamol product |
| 20240415-36-2_063 | fiz_d2 | blank | Paracetamol product |
| 20240415-36-3_064 | fiz_d3 | blank | Paracetamol product |
| 20240415-36-3_065 | fiz_d3 | blank | Paracetamol product |
| 20240415-36-3_066 | fiz_d3 | blank | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-1-1-1_011 | fiz_e01 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-1-1-2_012 | fiz_e01 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-1-1-3_013 | fiz_e01 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-1-2-1_014 | fiz_e02 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-1-2-2_015 | fiz_e02 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-1-2-3_016 | fiz_e02 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-1-3-1_017 | fiz_e03 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-1-3-2_018 | fiz_e03 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-1-3-3_019 | fiz_e03 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-2-1-1_022 | fiz_e04 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-2-1-2_023 | fiz_e04 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-2-1-3_024 | fiz_e04 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-2-2-1_025 | fiz_e05 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-2-2-2_026 | fiz_e05 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-2-2-3_027 | fiz_e05 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-2-3-1_028 | fiz_e06 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-2-3-2_029 | fiz_e06 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-2-3-3_030 | fiz_e06 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-3-1-1_033 | fiz_e07 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-3-1-2_034 | fiz_e07 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-3-1-3_035 | fiz_e07 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-3-2-1_036 | fiz_e08 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-3-2-2_037 | fiz_e08 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-3-2-3_038 | fiz_e08 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-3-3-1_039 | fiz_e09 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-3-3-2_040 | fiz_e09 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-3-3-3_041 | fiz_e09 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-4-1-1_044 | fiz_e10 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-4-1-2_045 | fiz_e10 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-4-1-3_046 | fiz_e10 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-4-2-1_047 | fiz_e11 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-4-2-2_048 | fiz_e11 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-4-3-1_050 | fiz_e11 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-4-3-2_051 | fiz_e12 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-4-3-3_052 | fiz_e12 | blank_e | Paracetamol product |
| 20240516_Paracetamol_5162024_Paracetamol-Coffein-Aspirin_12-42-3_049 | fiz_e12 | blank_e | Paracetamol product |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-41-1_113 | par_1 | blank | Paracetamol standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-41-1_114 | par_1 | blank | Paracetamol standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-41-1_115 | par_1 | blank | Paracetamol standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-41-2_116 | par_2 | blank | Paracetamol standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-41-2_117 | par_2 | blank | Paracetamol standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-41-2_118 | par_2 | blank | Paracetamol standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-41-3_119 | par_3 | blank | Paracetamol standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-41-3_120 | par_3 | blank | Paracetamol standard |
| 20240415_Paracetamol_4152024_Paracetamol-Coffein-Aspirin_20240415-41-3_121 | par_3 | blank | Paracetamol standard |
ms_workflow_settings <- list(
MassSpecSettings_BinSpectra_StreamFind(bins = list("rt" = 5, "mz" = 2), refBinAnalysis = 1),
MassSpecSettings_NormalizeSpectra_minmax(),
MassSpecSettings_AverageSpectra_StreamFind(),
MassSpecSettings_SubtractBlankSpectra_StreamFind(negativeToZero = TRUE),
MassSpecSettings_NormalizeSpectra_blockweight()
)
ms$add_settings(ms_workflow_settings)
ms$print_workflow()
Workflow
1: BinSpectra (StreamFind)
2: NormalizeSpectra (minmax)
3: AverageSpectra (StreamFind)
4: SubtractBlankSpectra (StreamFind)
5: NormalizeSpectra (blockweight)
ms$run_workflow()
ms$plot_spectra()Pre-processed spectra for each analysis. Retention time dimension in he created bins is collapsed for each mass-to-charge ratio
ms$get_spectra()[1:10, ] analysis replicate polarity rt mz intensity bins blank
<char> <char> <int> <num> <num> <num> <char> <num>
1: ass_1 ass_1 1 0 56 7.223730e-04 0-56 0.4806362
2: ass_1 ass_1 1 5 56 0.000000e+00 5-56 0.2783423
3: ass_1 ass_1 1 10 56 8.938603e-04 10-56 0.2396827
4: ass_1 ass_1 1 15 56 7.981044e-04 15-56 0.4825239
5: ass_1 ass_1 1 20 56 5.882682e-04 20-56 0.5144360
6: ass_1 ass_1 1 25 56 7.743778e-04 25-56 0.6433077
7: ass_1 ass_1 1 30 56 4.271764e-04 30-56 0.9462764
8: ass_1 ass_1 1 35 56 0.000000e+00 35-56 0.8639968
9: ass_1 ass_1 1 40 56 0.000000e+00 40-56 0.5787367
10: ass_1 ass_1 1 45 56 3.947558e-05 45-56 0.5303741Raman
| analysis | replicate | blank | description |
|---|---|---|---|
| 240411_1mgmLASS_200mW_OD3_2s_36n_pos1_r1 | ass_1 | blank | Aspirin standard |
| 240411_1mgmLASS_200mW_OD3_2s_36n_pos1_r2 | ass_1 | blank | Aspirin standard |
| 240411_1mgmLASS_200mW_OD3_2s_36n_pos1_r3 | ass_1 | blank | Aspirin standard |
| 240411_1mgmLASS_200mW_OD3_2s_36n_pos2_r1 | ass_2 | blank | Aspirin standard |
| 240411_1mgmLASS_200mW_OD3_2s_36n_pos2_r2 | ass_2 | blank | Aspirin standard |
| 240411_1mgmLASS_200mW_OD3_2s_36n_pos2_r3 | ass_2 | blank | Aspirin standard |
| 240411_1mgmLASS_200mW_OD3_2s_36n_pos3_r1 | ass_3 | blank | Aspirin standard |
| 240411_1mgmLASS_200mW_OD3_2s_36n_pos3_r2 | ass_3 | blank | Aspirin standard |
| 240411_1mgmLASS_200mW_OD3_2s_36n_pos3_r3 | ass_3 | blank | Aspirin standard |
| 240411_blank_200mW_OD3_2s_36n_pos1_r1 | blank | blank | Reference background |
| 240411_blank_200mW_OD3_2s_36n_pos1_r2 | blank | blank | Reference background |
| 240411_blank_200mW_OD3_2s_36n_pos1_r3 | blank | blank | Reference background |
| 240411_blank_200mW_OD3_2s_36n_pos2_r1 | blank | blank | Reference background |
| 240411_blank_200mW_OD3_2s_36n_pos2_r2 | blank | blank | Reference background |
| 240411_blank_200mW_OD3_2s_36n_pos2_r3 | blank | blank | Reference background |
| 240411_blank_200mW_OD3_2s_36n_pos3_r1 | blank | blank | Reference background |
| 240411_blank_200mW_OD3_2s_36n_pos3_r2 | blank | blank | Reference background |
| 240411_blank_200mW_OD3_2s_36n_pos3_r3 | blank | blank | Reference background |
| 240412_BlankH2O_200mW_OD3_2s_36n_pos1_r1 | blank_e | blank_e | Reference background |
| 240412_BlankH2O_200mW_OD3_2s_36n_pos1_r2 | blank_e | blank_e | Reference background |
| 240412_BlankH2O_200mW_OD3_2s_36n_pos1_r3 | blank_e | blank_e | Reference background |
| 240412_BlankH2O_200mW_OD3_2s_36n_pos2_r1 | blank_e | blank_e | Reference background |
| 240412_BlankH2O_200mW_OD3_2s_36n_pos2_r2 | blank_e | blank_e | Reference background |
| 240412_BlankH2O_200mW_OD3_2s_36n_pos2_r3 | blank_e | blank_e | Reference background |
| 240412_BlankH2O_200mW_OD3_2s_36n_pos3_r1 | blank_e | blank_e | Reference background |
| 240412_BlankH2O_200mW_OD3_2s_36n_pos3_r2 | blank_e | blank_e | Reference background |
| 240412_BlankH2O_200mW_OD3_2s_36n_pos3_r3 | blank_e | blank_e | Reference background |
| 240411_1mgmLCoffein_200mW_OD3_2s_36n_pos1_r1 | caff_1 | blank | Caffeine standard |
| 240411_1mgmLCoffein_200mW_OD3_2s_36n_pos1_r2 | caff_1 | blank | Caffeine standard |
| 240411_1mgmLCoffein_200mW_OD3_2s_36n_pos1_r3 | caff_1 | blank | Caffeine standard |
| 240411_1mgmLCoffein_200mW_OD3_2s_36n_pos2_r1 | caff_2 | blank | Caffeine standard |
| 240411_1mgmLCoffein_200mW_OD3_2s_36n_pos2_r2 | caff_2 | blank | Caffeine standard |
| 240411_1mgmLCoffein_200mW_OD3_2s_36n_pos2_r3 | caff_2 | blank | Caffeine standard |
| 240411_1mgmLCoffein_200mW_OD3_2s_36n_pos3_r1 | caff_3 | blank | Caffeine standard |
| 240411_1mgmLCoffein_200mW_OD3_2s_36n_pos3_r2 | caff_3 | blank | Caffeine standard |
| 240411_1mgmLCoffein_200mW_OD3_2s_36n_pos3_r3 | caff_3 | blank | Caffeine standard |
| 240411_Fizamol-11gealtert_200mW_OD3_2s_36n_pos1_r1 | fiz_a1 | blank | Paracetamol product |
| 240411_Fizamol-11gealtert_200mW_OD3_2s_36n_pos1_r2 | fiz_a1 | blank | Paracetamol product |
| 240411_Fizamol-11gealtert_200mW_OD3_2s_36n_pos1_r3 | fiz_a1 | blank | Paracetamol product |
| 240411_Fizamol-11gealtert_200mW_OD3_2s_36n_pos2_r1 | fiz_a2 | blank | Paracetamol product |
| 240411_Fizamol-11gealtert_200mW_OD3_2s_36n_pos2_r2 | fiz_a2 | blank | Paracetamol product |
| 240411_Fizamol-11gealtert_200mW_OD3_2s_36n_pos2_r3 | fiz_a2 | blank | Paracetamol product |
| 240411_Fizamol-11gealtert_200mW_OD3_2s_36n_pos3_r1 | fiz_a3 | blank | Paracetamol product |
| 240411_Fizamol-11gealtert_200mW_OD3_2s_36n_pos3_r2 | fiz_a3 | blank | Paracetamol product |
| 240411_Fizamol-11gealtert_200mW_OD3_2s_36n_pos3_r3 | fiz_a3 | blank | Paracetamol product |
| 240412_Fizamol-11+2prozentEssig_200mW_OD3_2s_36n_pos1_r1 | fiz_acid1 | blank | Paracetamol with acetic acid |
| 240412_Fizamol-11+2prozentEssig_200mW_OD3_2s_36n_pos1_r2 | fiz_acid1 | blank | Paracetamol with acetic acid |
| 240412_Fizamol-11+2prozentEssig_200mW_OD3_2s_36n_pos1_r3 | fiz_acid1 | blank | Paracetamol with acetic acid |
| 240412_Fizamol-11+2prozentEssig_200mW_OD3_2s_36n_pos2_r1 | fiz_acid2 | blank | Paracetamol with acetic acid |
| 240412_Fizamol-11+2prozentEssig_200mW_OD3_2s_36n_pos2_r2 | fiz_acid2 | blank | Paracetamol with acetic acid |
| 240412_Fizamol-11+2prozentEssig_200mW_OD3_2s_36n_pos2_r3 | fiz_acid2 | blank | Paracetamol with acetic acid |
| 240412_Fizamol-11+2prozentEssig_200mW_OD3_2s_36n_pos3_r1 | fiz_acid3 | blank | Paracetamol with acetic acid |
| 240412_Fizamol-11+2prozentEssig_200mW_OD3_2s_36n_pos3_r2 | fiz_acid3 | blank | Paracetamol with acetic acid |
| 240412_Fizamol-11+2prozentEssig_200mW_OD3_2s_36n_pos3_r3 | fiz_acid3 | blank | Paracetamol with acetic acid |
| 240411_Fizamol-11+1mgmLASS2_200mW_OD3_2s_36n_pos1_r1 | fiz_ass1 | blank | Paracetamol with aspirin |
| 240411_Fizamol-11+1mgmLASS2_200mW_OD3_2s_36n_pos1_r2 | fiz_ass1 | blank | Paracetamol with aspirin |
| 240411_Fizamol-11+1mgmLASS2_200mW_OD3_2s_36n_pos1_r3 | fiz_ass1 | blank | Paracetamol with aspirin |
| 240411_Fizamol-11+1mgmLASS2_200mW_OD3_2s_36n_pos2_r1 | fiz_ass2 | blank | Paracetamol with aspirin |
| 240411_Fizamol-11+1mgmLASS2_200mW_OD3_2s_36n_pos2_r2 | fiz_ass2 | blank | Paracetamol with aspirin |
| 240411_Fizamol-11+1mgmLASS2_200mW_OD3_2s_36n_pos2_r3 | fiz_ass2 | blank | Paracetamol with aspirin |
| 240411_Fizamol-11+1mgmLASS2_200mW_OD3_2s_36n_pos3_r1 | fiz_ass3 | blank | Paracetamol with aspirin |
| 240411_Fizamol-11+1mgmLASS2_200mW_OD3_2s_36n_pos3_r2 | fiz_ass3 | blank | Paracetamol with aspirin |
| 240411_Fizamol-11+1mgmLASS2_200mW_OD3_2s_36n_pos3_r3 | fiz_ass3 | blank | Paracetamol with aspirin |
| 240411_Fizamol-12gealtert_200mW_OD3_2s_36n_pos1_r1 | fiz_b1 | blank | Paracetamol product |
| 240411_Fizamol-12gealtert_200mW_OD3_2s_36n_pos1_r2 | fiz_b1 | blank | Paracetamol product |
| 240411_Fizamol-12gealtert_200mW_OD3_2s_36n_pos1_r3 | fiz_b1 | blank | Paracetamol product |
| 240411_Fizamol-12gealtert_200mW_OD3_2s_36n_pos2_r1 | fiz_b2 | blank | Paracetamol product |
| 240411_Fizamol-12gealtert_200mW_OD3_2s_36n_pos2_r2 | fiz_b2 | blank | Paracetamol product |
| 240411_Fizamol-12gealtert_200mW_OD3_2s_36n_pos2_r3 | fiz_b2 | blank | Paracetamol product |
| 240411_Fizamol-12gealtert_200mW_OD3_2s_36n_pos3_r1 | fiz_b3 | blank | Paracetamol product |
| 240411_Fizamol-12gealtert_200mW_OD3_2s_36n_pos3_r2 | fiz_b3 | blank | Paracetamol product |
| 240411_Fizamol-12gealtert_200mW_OD3_2s_36n_pos3_r3 | fiz_b3 | blank | Paracetamol product |
| 240411_Fizamol-11_200mW_OD3_2s_36n_pos1_r1 | fiz_c1 | blank | Paracetamol product |
| 240411_Fizamol-11_200mW_OD3_2s_36n_pos1_r2 | fiz_c1 | blank | Paracetamol product |
| 240411_Fizamol-11_200mW_OD3_2s_36n_pos1_r3 | fiz_c1 | blank | Paracetamol product |
| 240411_Fizamol-11_200mW_OD3_2s_36n_pos2_r1 | fiz_c2 | blank | Paracetamol product |
| 240411_Fizamol-11_200mW_OD3_2s_36n_pos2_r2 | fiz_c2 | blank | Paracetamol product |
| 240411_Fizamol-11_200mW_OD3_2s_36n_pos2_r3 | fiz_c2 | blank | Paracetamol product |
| 240411_Fizamol-11_200mW_OD3_2s_36n_pos3_r1 | fiz_c3 | blank | Paracetamol product |
| 240411_Fizamol-11_200mW_OD3_2s_36n_pos3_r2 | fiz_c3 | blank | Paracetamol product |
| 240411_Fizamol-11_200mW_OD3_2s_36n_pos3_r3 | fiz_c3 | blank | Paracetamol product |
| 240411_Fizamol-11+1mgmLcoffein_200mW_OD3_2s_36n_pos1_r1 | fiz_caff1 | blank | Paracetamol with caffeine |
| 240411_Fizamol-11+1mgmLcoffein_200mW_OD3_2s_36n_pos1_r2 | fiz_caff1 | blank | Paracetamol with caffeine |
| 240411_Fizamol-11+1mgmLcoffein_200mW_OD3_2s_36n_pos1_r3 | fiz_caff1 | blank | Paracetamol with caffeine |
| 240411_Fizamol-11+1mgmLcoffein_200mW_OD3_2s_36n_pos2_r1 | fiz_caff2 | blank | Paracetamol with caffeine |
| 240411_Fizamol-11+1mgmLcoffein_200mW_OD3_2s_36n_pos2_r2 | fiz_caff2 | blank | Paracetamol with caffeine |
| 240411_Fizamol-11+1mgmLcoffein_200mW_OD3_2s_36n_pos2_r3 | fiz_caff2 | blank | Paracetamol with caffeine |
| 240411_Fizamol-11+1mgmLcoffein_200mW_OD3_2s_36n_pos3_r1 | fiz_caff3 | blank | Paracetamol with caffeine |
| 240411_Fizamol-11+1mgmLcoffein_200mW_OD3_2s_36n_pos3_r2 | fiz_caff3 | blank | Paracetamol with caffeine |
| 240411_Fizamol-11+1mgmLcoffein_200mW_OD3_2s_36n_pos3_r3 | fiz_caff3 | blank | Paracetamol with caffeine |
| 240411_Fizamol-12_200mW_OD3_2s_36n_pos1_r1 | fiz_d1 | blank | Paracetamol product |
| 240411_Fizamol-12_200mW_OD3_2s_36n_pos1_r2 | fiz_d1 | blank | Paracetamol product |
| 240411_Fizamol-12_200mW_OD3_2s_36n_pos1_r3 | fiz_d1 | blank | Paracetamol product |
| 240411_Fizamol-12_200mW_OD3_2s_36n_pos2_r1 | fiz_d2 | blank | Paracetamol product |
| 240411_Fizamol-12_200mW_OD3_2s_36n_pos2_r2 | fiz_d2 | blank | Paracetamol product |
| 240411_Fizamol-12_200mW_OD3_2s_36n_pos2_r3 | fiz_d2 | blank | Paracetamol product |
| 240411_Fizamol-12_200mW_OD3_2s_36n_pos3_r1 | fiz_d3 | blank | Paracetamol product |
| 240411_Fizamol-12_200mW_OD3_2s_36n_pos3_r2 | fiz_d3 | blank | Paracetamol product |
| 240411_Fizamol-12_200mW_OD3_2s_36n_pos3_r3 | fiz_d3 | blank | Paracetamol product |
| 240412_Fizamol12-1_200mW_OD3_2s_36n_pos1_r1 | fiz_e01 | blank_e | Paracetamol product |
| 240412_Fizamol12-1_200mW_OD3_2s_36n_pos1_r2 | fiz_e01 | blank_e | Paracetamol product |
| 240412_Fizamol12-1_200mW_OD3_2s_36n_pos1_r3 | fiz_e01 | blank_e | Paracetamol product |
| 240412_Fizamol12-1_200mW_OD3_2s_36n_pos2_r1 | fiz_e02 | blank_e | Paracetamol product |
| 240412_Fizamol12-1_200mW_OD3_2s_36n_pos2_r2 | fiz_e02 | blank_e | Paracetamol product |
| 240412_Fizamol12-1_200mW_OD3_2s_36n_pos2_r3 | fiz_e02 | blank_e | Paracetamol product |
| 240412_Fizamol12-1_200mW_OD3_2s_36n_pos3_r1 | fiz_e03 | blank_e | Paracetamol product |
| 240412_Fizamol12-1_200mW_OD3_2s_36n_pos3_r2 | fiz_e03 | blank_e | Paracetamol product |
| 240412_Fizamol12-1_200mW_OD3_2s_36n_pos3_r3 | fiz_e03 | blank_e | Paracetamol product |
| 240412_Fizamol12-2_200mW_OD3_2s_36n_pos1_r1 | fiz_e04 | blank_e | Paracetamol product |
| 240412_Fizamol12-2_200mW_OD3_2s_36n_pos1_r2 | fiz_e04 | blank_e | Paracetamol product |
| 240412_Fizamol12-2_200mW_OD3_2s_36n_pos1_r3 | fiz_e04 | blank_e | Paracetamol product |
| 240412_Fizamol12-2_200mW_OD3_2s_36n_pos2_r1 | fiz_e05 | blank_e | Paracetamol product |
| 240412_Fizamol12-2_200mW_OD3_2s_36n_pos2_r2 | fiz_e05 | blank_e | Paracetamol product |
| 240412_Fizamol12-2_200mW_OD3_2s_36n_pos2_r3 | fiz_e05 | blank_e | Paracetamol product |
| 240412_Fizamol12-2_200mW_OD3_2s_36n_pos3_r1 | fiz_e06 | blank_e | Paracetamol product |
| 240412_Fizamol12-2_200mW_OD3_2s_36n_pos3_r2 | fiz_e06 | blank_e | Paracetamol product |
| 240412_Fizamol12-2_200mW_OD3_2s_36n_pos3_r3 | fiz_e06 | blank_e | Paracetamol product |
| 240412_Fizamol12-3_200mW_OD3_2s_36n_pos1_r1 | fiz_e07 | blank_e | Paracetamol product |
| 240412_Fizamol12-3_200mW_OD3_2s_36n_pos1_r2 | fiz_e07 | blank_e | Paracetamol product |
| 240412_Fizamol12-3_200mW_OD3_2s_36n_pos1_r3 | fiz_e07 | blank_e | Paracetamol product |
| 240412_Fizamol12-3_200mW_OD3_2s_36n_pos2_r1 | fiz_e08 | blank_e | Paracetamol product |
| 240412_Fizamol12-3_200mW_OD3_2s_36n_pos2_r2 | fiz_e08 | blank_e | Paracetamol product |
| 240412_Fizamol12-3_200mW_OD3_2s_36n_pos2_r3 | fiz_e08 | blank_e | Paracetamol product |
| 240412_Fizamol12-3_200mW_OD3_2s_36n_pos3_r1 | fiz_e09 | blank_e | Paracetamol product |
| 240412_Fizamol12-3_200mW_OD3_2s_36n_pos3_r2 | fiz_e09 | blank_e | Paracetamol product |
| 240412_Fizamol12-3_200mW_OD3_2s_36n_pos3_r3 | fiz_e09 | blank_e | Paracetamol product |
| 240412_Fizamol12-4_200mW_OD3_2s_36n_pos1_r1 | fiz_e10 | blank_e | Paracetamol product |
| 240412_Fizamol12-4_200mW_OD3_2s_36n_pos1_r2 | fiz_e10 | blank_e | Paracetamol product |
| 240412_Fizamol12-4_200mW_OD3_2s_36n_pos1_r3 | fiz_e10 | blank_e | Paracetamol product |
| 240412_Fizamol12-4_200mW_OD3_2s_36n_pos2_r1 | fiz_e11 | blank_e | Paracetamol product |
| 240412_Fizamol12-4_200mW_OD3_2s_36n_pos2_r2 | fiz_e11 | blank_e | Paracetamol product |
| 240412_Fizamol12-4_200mW_OD3_2s_36n_pos2_r3 | fiz_e11 | blank_e | Paracetamol product |
| 240412_Fizamol12-4_200mW_OD3_2s_36n_pos3_r1 | fiz_e12 | blank_e | Paracetamol product |
| 240412_Fizamol12-4_200mW_OD3_2s_36n_pos3_r2 | fiz_e12 | blank_e | Paracetamol product |
| 240412_Fizamol12-4_200mW_OD3_2s_36n_pos3_r3 | fiz_e12 | blank_e | Paracetamol product |
| 240412_Paracetamol-10mgmL_200mW_OD3_2s_36n_pos1_r1 | par_1 | blank | Paracetamol standard |
| 240412_Paracetamol-10mgmL_200mW_OD3_2s_36n_pos1_r2 | par_1 | blank | Paracetamol standard |
| 240412_Paracetamol-10mgmL_200mW_OD3_2s_36n_pos1_r3 | par_1 | blank | Paracetamol standard |
| 240412_Paracetamol-10mgmL_200mW_OD3_2s_36n_pos4_r1 | par_2 | blank | Paracetamol standard |
| 240412_Paracetamol-10mgmL_200mW_OD3_2s_36n_pos4_r2 | par_2 | blank | Paracetamol standard |
| 240412_Paracetamol-10mgmL_200mW_OD3_2s_36n_pos4_r3 | par_2 | blank | Paracetamol standard |
| 240412_Paracetamol-10mgmL_200mW_OD3_2s_36n_pos3_r1 | par_3 | blank | Paracetamol standard |
| 240412_Paracetamol-10mgmL_200mW_OD3_2s_36n_pos3_r2 | par_3 | blank | Paracetamol standard |
| 240412_Paracetamol-10mgmL_200mW_OD3_2s_36n_pos3_r3 | par_3 | blank | Paracetamol standard |
raman_workflow_settings <- list(
RamanSettings_AverageSpectra_StreamFind(),
RamanSettings_SubtractBlankSpectra_StreamFind(),
RamanSettings_SmoothSpectra_savgol(fl = 21, forder = 4, dorder = 0),
RamanSettings_DeleteSpectraSection_StreamFind(list("shift" = c(-100, 350))),
RamanSettings_DeleteSpectraSection_StreamFind(list("shift" = c(1800, 2500))),
RamanSettings_CorrectSpectraBaseline_airpls(lambda = 45, differences = 1, itermax = 30),
RamanSettings_NormalizeSpectra_minmax(),
RamanSettings_NormalizeSpectra_blockweight()
)
raman$add_settings(raman_workflow_settings)
raman$print_workflow()
Workflow
1: AverageSpectra (StreamFind)
2: SubtractBlankSpectra (StreamFind)
3: SmoothSpectra (savgol)
4: DeleteSpectraSection (StreamFind)
5: DeleteSpectraSection (StreamFind)
6: CorrectSpectraBaseline (airpls)
7: NormalizeSpectra (minmax)
8: NormalizeSpectra (blockweight)
raman$run_workflow()
raman$plot_spectra()Pre-processed Raman spectra for analysis replicate.
raman$get_spectra()[1:10, ] analysis replicate shift intensity blank baseline raw
<char> <char> <num> <num> <num> <num> <num>
1: ass_1 ass_1 350.3578 0.000000000 47160.00 9148.558 8954.093
2: ass_1 ass_1 353.3808 0.000000000 47245.78 9152.879 9038.648
3: ass_1 ass_1 356.4008 0.000000000 47054.00 9159.943 9080.207
4: ass_1 ass_1 359.4177 0.000000000 46981.00 9168.890 9106.521
5: ass_1 ass_1 362.4336 0.000000000 47145.78 9179.299 9176.353
6: ass_1 ass_1 365.4485 0.002669691 46989.00 9189.776 9229.674
7: ass_1 ass_1 368.4604 0.004423876 46941.89 9199.360 9265.474
8: ass_1 ass_1 371.4691 0.005247175 46765.00 9208.944 9287.362
9: ass_1 ass_1 374.4769 0.006030455 47019.67 9218.528 9308.653
10: ass_1 ass_1 377.4838 0.007325727 46920.44 9228.112 9337.595Fusion
ms_spec_mat <- ms$get_spectra_matrix()
raman_spec_mat <- raman$get_spectra_matrix()
fused_mat <- cbind(ms_spec_mat, raman_spec_mat)
# The matrix is mean centered using the mdatools R package
fused_mat <- mdatools::prep.autoscale(fused_mat, center = TRUE, scale = FALSE)Statistic analysis
Unsupervised
Principle Components Analysis (PCA) can be used for unsupervised evaluation, meaning that we assume that the main (or train) set of analyses are of good quality and we test new analyses to evaluate the proximity to the main set. One could say that it is a supervised approach but in the sense that we assume the quality of the main set. Yet, we are not classifying the possible deviation of the new analyses which would be a clear supervised approach, as we show in the next sub chapter.
[1] "fiz_a1" "fiz_a2" "fiz_a3" "fiz_b1" "fiz_b2" "fiz_b3" "fiz_c1"
[8] "fiz_c2" "fiz_c3" "fiz_d1" "fiz_d2" "fiz_d3" "fiz_e01" "fiz_e02"
[15] "fiz_e03" "fiz_e04" "fiz_e05" "fiz_e06" [1] "ass_1" "ass_2" "ass_3" "caff_1" "caff_2" "caff_3"
[7] "fiz_acid1" "fiz_acid2" "fiz_acid3" "fiz_ass1" "fiz_ass2" "fiz_ass3"
[13] "fiz_caff1" "fiz_caff2" "fiz_caff3" "fiz_e07" "fiz_e08" "fiz_e09"
[19] "fiz_e10" "fiz_e11" "fiz_e12" "par_1" "par_2" "par_3"
stat <- StatisticEngine$new(data = main_set)
# The alpha and gamma parameters are used to evaluate the model limits
stat$process(StatisticSettings_MakeModel_pca_mdatools(ncomp = 3, alpha = 1E-4, gamma = 1E-5))
# Original plot from mdatools R package
plot(stat$model)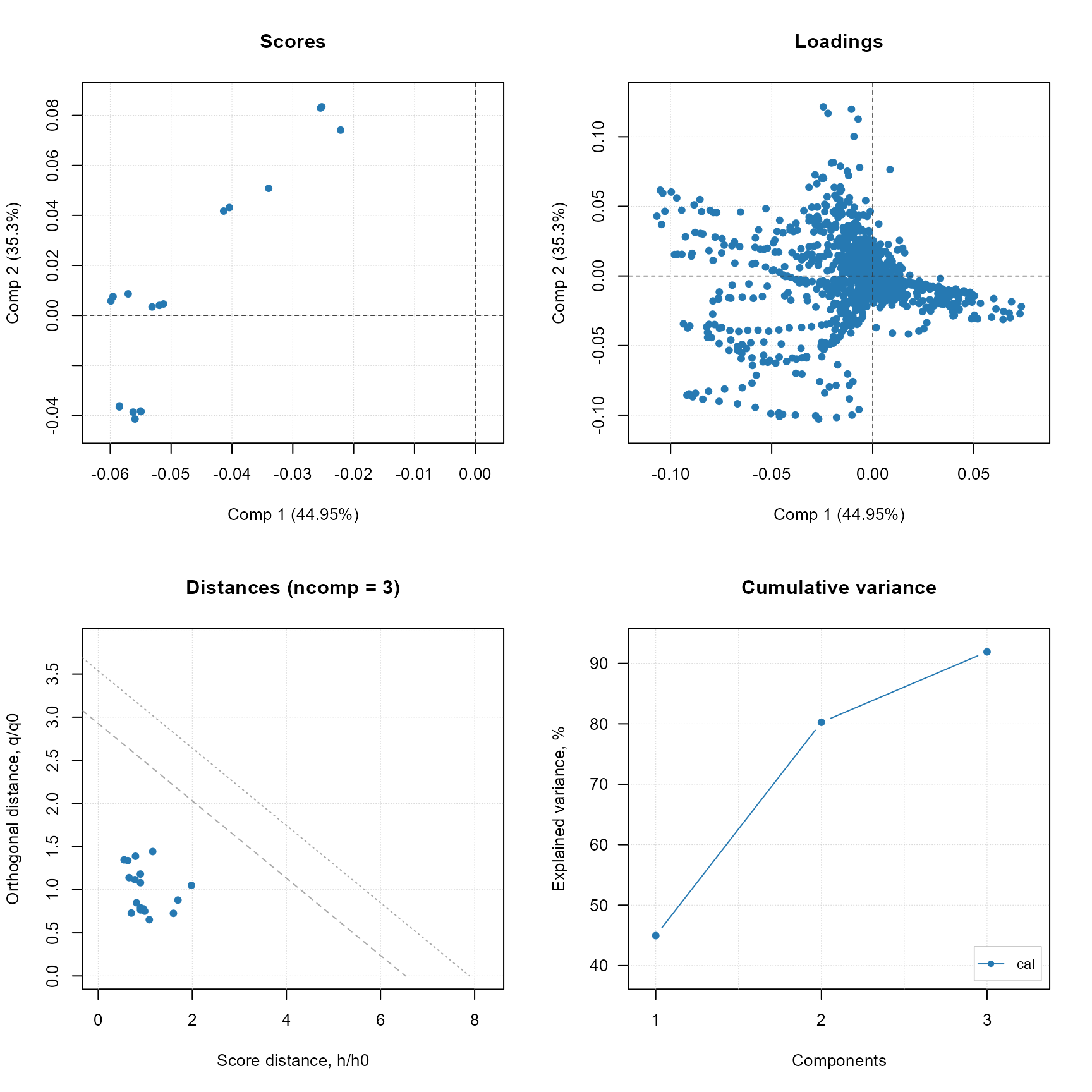
PCA model (native plot from mdatools R package).
stat$predict(test_set)
stat$plot_predicted_distances()PCA model prediction distances. For more information about the distances plese consult the mdatools guide in https://mdatools.com/docs/pca--distances-and-limits.html.
Supervised (classification)
The k-nearest neighbors (KNN) can be used for classification of analyses based on pre-classified training analyses.
train_set <- fused_mat[unique(c(grep("1", rownames(fused_mat)), grep("2", rownames(fused_mat)))), ]
train_set <- train_set[order(rownames(train_set)), ]
rownames(train_set) [1] "ass_1" "ass_2" "caff_1" "caff_2" "fiz_a1" "fiz_a2"
[7] "fiz_acid1" "fiz_acid2" "fiz_ass1" "fiz_ass2" "fiz_b1" "fiz_b2"
[13] "fiz_c1" "fiz_c2" "fiz_caff1" "fiz_caff2" "fiz_d1" "fiz_d2"
[19] "fiz_e01" "fiz_e02" "fiz_e10" "fiz_e11" "fiz_e12" "par_1"
[25] "par_2" [1] "ass_3" "caff_3" "fiz_a3" "fiz_acid3" "fiz_ass3" "fiz_b3"
[7] "fiz_c3" "fiz_caff3" "fiz_d3" "fiz_e03" "fiz_e04" "fiz_e05"
[13] "fiz_e06" "fiz_e07" "fiz_e08" "fiz_e09" "par_3"
labels <- c(rep("ass", 2), rep("caff", 2), rep("fiz", 2), rep("fiz_acid", 2), rep("fiz_ass", 2),
rep("fiz", 2), rep("fiz", 2), rep("fiz_caff", 2), rep("fiz", 7), rep("par", 2))
stat2 <- StatisticEngine$new(data = train_set)
stat2$add_classes(labels)
stat2$process(StatisticSettings_PrepareClassification_knn(k = 3))
stat2$classify(test_set)
stat2$classification_results analysis class probability
<char> <fctr> <num>
1: ass_3 ass 0.7
2: caff_3 caff 0.7
3: fiz_a3 fiz 1.0
4: fiz_acid3 fiz_acid 0.7
5: fiz_ass3 fiz_ass 0.7
6: fiz_b3 fiz 0.7
7: fiz_c3 fiz 1.0
8: fiz_caff3 fiz_caff 0.7
9: fiz_d3 fiz 1.0
10: fiz_e03 fiz 1.0
11: fiz_e04 fiz 1.0
12: fiz_e05 fiz 1.0
13: fiz_e06 fiz 1.0
14: fiz_e07 fiz 1.0
15: fiz_e08 fiz 1.0
16: fiz_e09 fiz 1.0
17: par_3 par 0.7Only MS
ms_spec_mat <- mdatools::prep.autoscale(ms_spec_mat, center = TRUE, scale = FALSE)
main_set_ms <- ms_spec_mat[c(7:9, 16:21, 25:33), ]
test_set_ms <- ms_spec_mat[c(1:6, 10:15, 22:24, 34:42), ]
stat_ms <- StatisticEngine$new(data = main_set_ms)
stat_ms$process(StatisticSettings_MakeModel_pca_mdatools(ncomp = 3, alpha = 1E-4, gamma = 1E-5))
plot(stat_ms$model)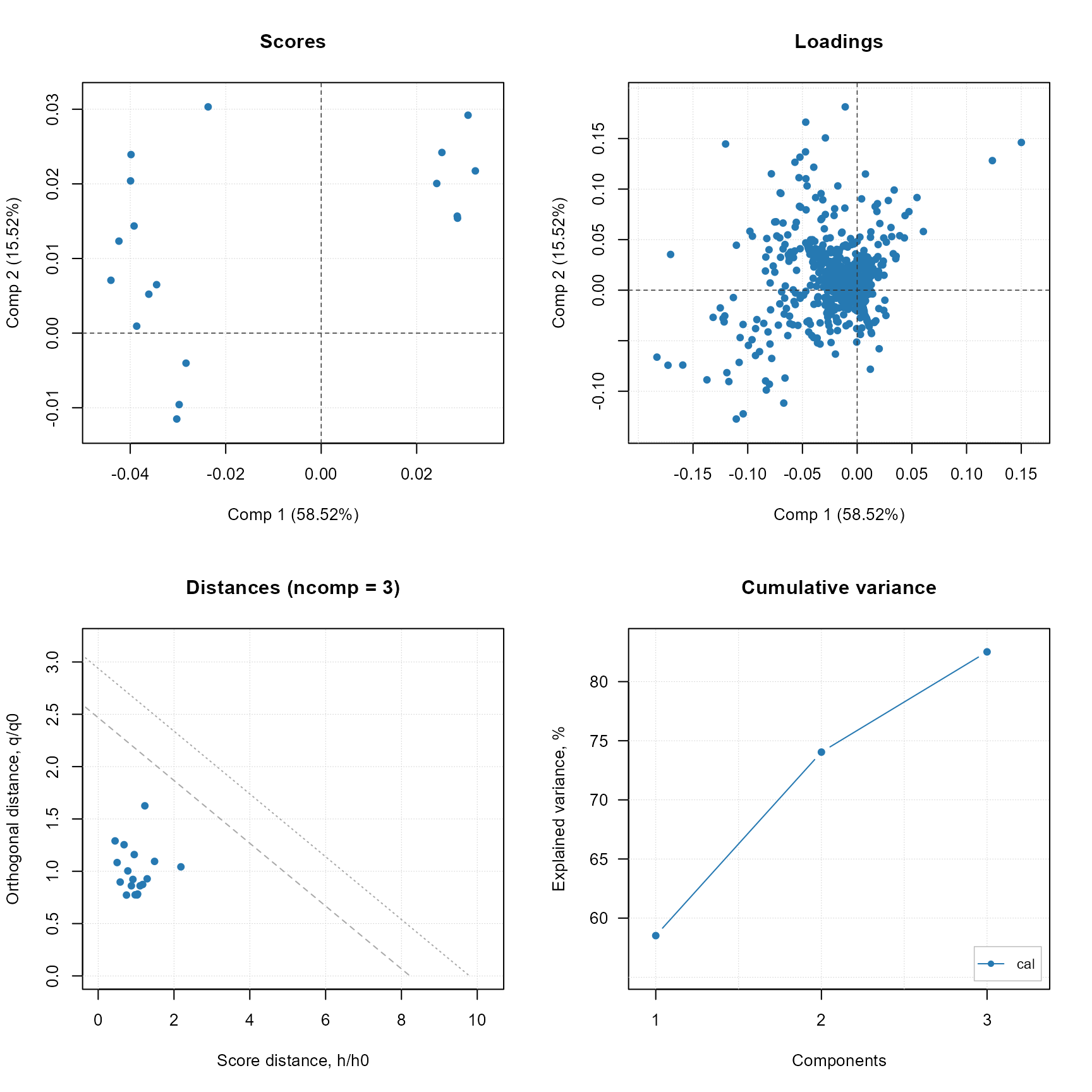
PCA model for MS data only (native plot from mdatools R package).
stat_ms$predict(test_set_ms)
stat_ms$plot_predicted_distances()PCA model prediction distances for only MS data.
Using MS data only for evaluation of quality deviations would not separate completely the analyses when compared to fused MS and Raman data. Thus, the contribution of Raman spectroscopy to explain structural deviations brings an advantage for routine quality evaluation of paracetamol and other pharmaceuticals.
Only Raman
raman_spec_mat <- mdatools::prep.autoscale(raman_spec_mat, center = TRUE, scale = FALSE)
main_set_raman <- raman_spec_mat[c(7:9, 16:21, 25:33), ]
test_set_raman <- raman_spec_mat[c(1:6, 10:15, 22:24, 34:42), ]
stat_raman <- StatisticEngine$new(data = main_set_raman)
stat_raman$process(StatisticSettings_MakeModel_pca_mdatools(ncomp = 3, alpha = 1E-4, gamma = 1E-5))
plot(stat_raman$model)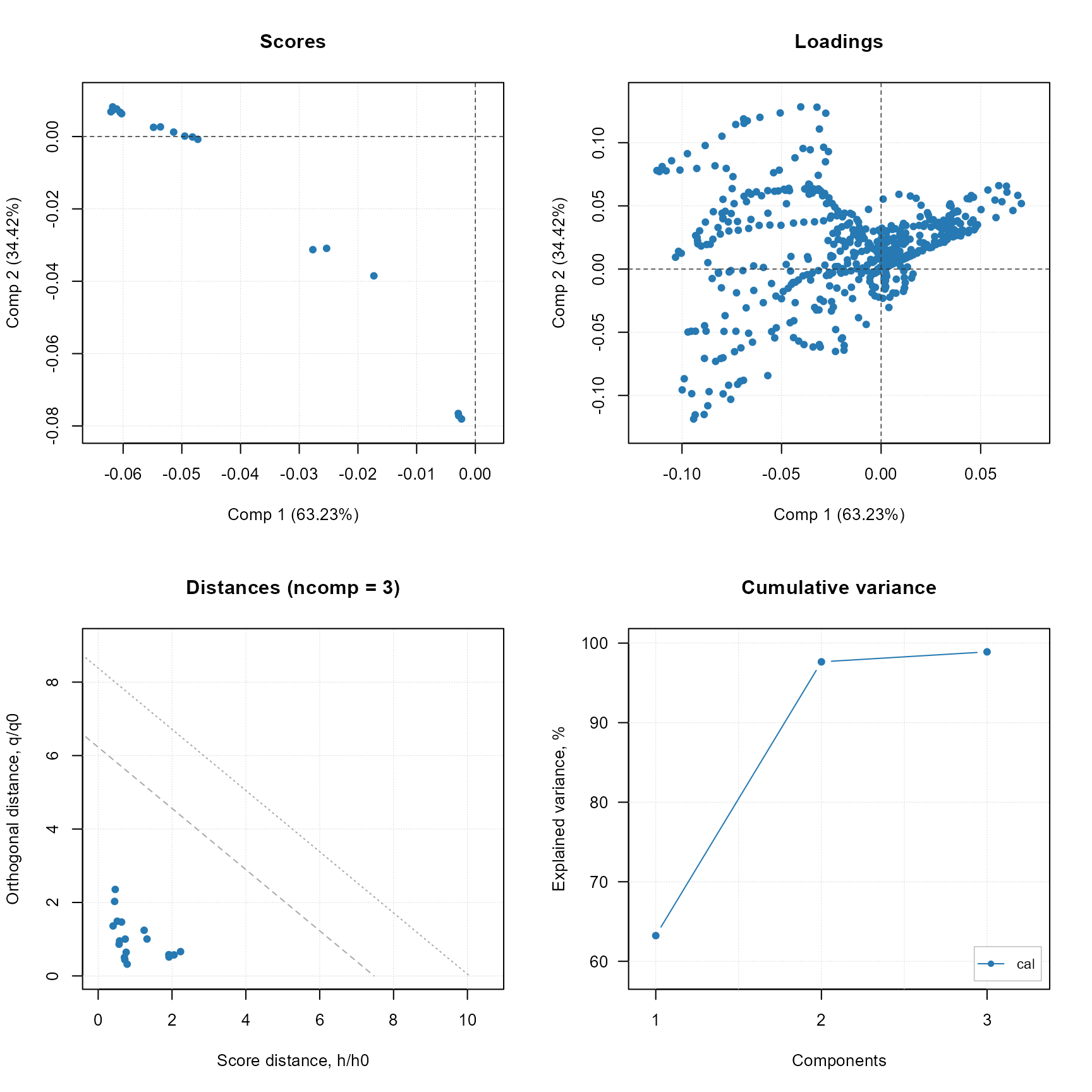
PCA model for Raman data only (native plot from mdatools R package).
stat_raman$predict(test_set_raman)
stat_raman$plot_predicted_distances()PCA model prediction distances for only Raman data.
In the case of only using Raman data for evaluation of quality deviations, the separation of the analyses is clearer than when using fused MS and Raman data. This is due to the fact that the Raman data is more sensitive to structural deviations in the analyses. The lack of aspirin and acetic acid signals in the MS data probably contributed to the less clear separation of the analyses when using only MS data. On the contrary, because the signal of caffeine (m/z 194 Da) was visible in the MS data, a separation of the caffeine contaminated analyses was obtained also for MS only data. MS data is more precise when the goal is identification of the active product ingredient (i.e, paracetamol) therefore, the fusion of MS and Raman data is recommended for quality evaluation of pharmaceuticals as both techniques are complementary.
Quantification with Raman
raman_quant$plot_spectra(colorBy = "replicates")Raw averaged Raman spectra for each analysis replicate.
raman_workflow_settings <- list(
RamanSettings_AverageSpectra_StreamFind(),
RamanSettings_SubtractBlankSpectra_StreamFind(),
RamanSettings_SmoothSpectra_savgol(fl = 21, forder = 4, dorder = 0),
RamanSettings_DeleteSpectraSection_StreamFind(list("shift" = c(-100, 350))),
RamanSettings_DeleteSpectraSection_StreamFind(list("shift" = c(1800, 2500))),
RamanSettings_CorrectSpectraBaseline_airpls(lambda = 45, differences = 1, itermax = 30)
)
raman_quant$add_settings(raman_workflow_settings)
raman_quant$print_workflow()
Workflow
1: AverageSpectra (StreamFind)
2: SubtractBlankSpectra (StreamFind)
3: SmoothSpectra (savgol)
4: DeleteSpectraSection (StreamFind)
5: DeleteSpectraSection (StreamFind)
6: CorrectSpectraBaseline (airpls)
raman_quant$run_workflow()
raman_quant$plot_spectra()Pre-processed Raman spectra for analysis replicate.
Applying MCR-ALS
The quantification of paracetamol from the Raman spectra is performed using the Multivariate Curve Resolution (MCR) with Alternating Least Squares (ALS) method via integration of the mdatools R package.
stat3 <- StatisticEngine$new(data = raman_quant$get_spectra_matrix())
stat3$add_concentrations(c(1, 2.5, 5.0, 7.5, 15.0, NA_real_)) # add concentrations, with NA for unknown
stat3$process(StatisticSettings_MakeModel_mcrals_mdatools(ncomp = 3))
summary(stat3$model)
Summary for MCR ALS case (class mcrals)
Constraints for spectra:
- none
Constraints for contributions:
- none
Expvar Cumexpvar
Comp 1 21.44 21.44
Comp 2 60.73 82.17
Comp 3 17.80 99.97
stat3$plot_model_resolved_spectra()Resolved spectra for each component and the spectra for each analysis replicate (i.e., each concentration).
stat3$plot_model_contributions()Contributions for each analysis replicate.
The quantify method can be used to estimate the
concentration of the paracetamol analyses with unknown concentration.
The quantification is applied to each component, returning a list with
the r square of the linear model and the estimated concentration as
data.table. Note that the explained variance for each
component is important to evaluate the calculated concentration. The
best estimation is from the component with the combined explained
variance and r square closer to 100 and 1, respectively.
stat3$quantify()$`Comp 1`
$`Comp 1`$r_squared
[1] 0.989119
$`Comp 1`$quantification
analysis mcr_value concentration
<char> <num> <num>
1: cali_010_mgml 2.833177 1.00000
2: cali_025_mgml 0.000000 2.50000
3: cali_050_mgml 196.055198 5.00000
4: cali_075_mgml 310.359083 7.50000
5: cali_150_mgml 711.390475 15.00000
6: par_3 888.288779 18.25022
$`Comp 2`
$`Comp 2`$r_squared
[1] 0.9735507
$`Comp 2`$quantification
analysis mcr_value concentration
<char> <num> <num>
1: cali_010_mgml 0.00000 1.000000
2: cali_025_mgml 16.13659 2.500000
3: cali_050_mgml 380.34701 5.000000
4: cali_075_mgml 566.07595 7.500000
5: cali_150_mgml 1846.37336 15.000000
6: par_3 815.00755 8.015858
$`Comp 3`
$`Comp 3`$r_squared
[1] 0.9632655
$`Comp 3`$quantification
analysis mcr_value concentration
<char> <num> <num>
1: cali_010_mgml 145.2248 1.000000
2: cali_025_mgml 340.0791 2.500000
3: cali_050_mgml 410.6169 5.000000
4: cali_075_mgml 642.4244 7.500000
5: cali_150_mgml 942.9335 15.000000
6: par_3 684.2477 9.514703