StreamFind General Introduction
Ricardo Cunha
cunha@iuta.de01 August, 2024
Source:vignettes/articles/framework.Rmd
framework.RmdThe StreamFind R package is a data processing workflow designer. Besides data processing, the platform can also be used for data management, visualization and reporting. This guide focuses on describing the general framework behind StreamFind. The StreamFind is centered around R6 classes, serving as data processing engines (used as metaphor) for different types of data (e.g. mass spectrometry (MS) and Raman spectroscopy data).
Data processing engines
Data processing engines are fundamentally reference classes with
methods to manage, process, visualize and report data within a project.
The CoreEngine is the parent class of all other data
specific engines (e.g. MassSpecEngine and
RamanEngine). As parent, the CoreEngine
manages the project information via the class
ProjectHeaders, registers the audit trail, and contains
uniform functions across child data dedicated engines (e.g. adding and
removing analyses from the project).
core <- CoreEngine$new()
core
CoreEngine
name NA
author NA
file NA
date 2024-08-01 10:06:20.248648
Workflow empty
Analyses empty
Results empty Note that when an empty CoreEngine is created, required
ProjectHeaders are created with name, author, path and
date. Yet, ProjectHeaders can be specified directly during
creation of the CoreEngine via the argument
headers or added to the engine as shown in
@ref(project-headers). The CoreEngine does not directly
handle data processing. Processing methods are data specific and
therefore, are used via the data dedicated engines. Yet, the framework
to manage the data processing workflow and the results are implemented
in the CoreEngine and are therefore, harmonized across
engines.
Project headers
The ProjectHeaders S3 class is meant to hold project
information that can be used to identify the engine when for example,
combining different engines, or to add extra information, such as
description, location, etc. The user can add any kind of attribute but
it must have length one and be named. Below, ProjectHeaders
are created and added to the CoreEngine.
headers <- ProjectHeaders(
name = "Project Example",
author = "Person Name",
description = "Example of project headers"
)
core$add_headers(headers)
core$get_headers()
ProjectHeaders
name: Project Example
author: Person Name
description: Example of project headers
file: NA
date: 2024-08-01 10:06:20.327951ProcessingSettings
Data processing workflows in StreamFind are assembled by combining
different processing methods in a specific order. Each processing method
uses a specific algorithm for processing/transforming data at a given
stage of the workflow. Thus, to harmonize the diversity of processing
methods and algorithms available, a general
ProcessingSettings is used (shown below). This way, the
ProcessingSettings objects are use as instructions to
assemble a data processing workflow within an engine.
ProcessingSettings
engine NA
call NA
algorithm NA
version 0.2.0
software NA
developer NA
contact NA
link NA
doi NA
parameters: empty A ProcessingSettings object must always have the engine
type, the call name of the processing method, the name of the algorithm
to be used, the origin software, the main developer name and contact as
well as a link to further information and the DOI, when available.
Lastly but not least, the parameters which is a flexible list of
conditions to apply the algorithm during data processing. As example,
ProcessingSettings for centroiding MS data using the
qCentroids from the qAlgorihtms and
for annotating features using a native algorithm from StreamFind are
shown below. Each ProcessingSettings object has a dedicated
constructor method with documentation to support the usage. Help pages
for processing methods can be obtained with the native R function
? or help().
ProcessingSettings
engine MassSpec
call CentroidSpectra
algorithm qCentroids
version 0.2.0
software qAlgorithms
developer Gerrit Renner
contact gerrit.renner@uni-due.de
link https://github.com/odea-project/qAlgorithms
doi https://doi.org/10.1007/s00216-022-04224-y
parameters:
- maxScale 5
- mode 1
ProcessingSettings
engine MassSpec
call AnnotateFeatures
algorithm StreamFind
version 0.2.0
software StreamFind
developer Ricardo Cunha
contact cunha@iuta.de
link https://odea-project.github.io/StreamFind
doi NA
parameters:
- maxIsotopes 5
- elements C H N O S Cl Br
- mode small molecules
- maxCharge 1
- rtWindowAlignment 0.3
- maxGaps 1 Saving and loading
The CoreEngine also holds the functionality to save the
project in the engine (as a SQLite file) and load it back. The
save() method saves the project and the load()
method loads it, as shown below.
file.exists(project_file_path)[1] TRUE
new_core <- CoreEngine$new()
new_core$load(project_file_path)
# The headers are has the core object although a new_core object was created with default headers
new_core$get_headers()
ProjectHeaders
name: Project Example
author: Person Name
description: Example of project headers
date: 2024-08-01 10:06:20.327951
file: C:/Users/apoli/Documents/github/StreamFind/vignettes/articles//project.sqliteData specific engines
As above mentioned, the CoreEngine does not handle data
processing directly. The data processing is delegated to child engines.
A simple example is given below by creating a child
RamanEngine and accessing the spectra from the analyses
(added as full paths to .asc files on disk). Note that the
workflow and results are still empty, as no data processing methods were
applied.
# Example raman .asc files
raman_ex_files <- StreamFindData::get_raman_file_paths()
raman <- RamanEngine$new(raman_ex_files)
raman
RamanEngine
name NA
author NA
file NA
date 2024-08-01 10:06:21.050755
Workflow empty
Analyses
analysis replicate blank spectra
<char> <char> <char> <num>
1: raman_Bevacizumab_11731 raman_Bevacizumab_11731 <NA> 1
2: raman_Bevacizumab_11732 raman_Bevacizumab_11732 <NA> 1
3: raman_Bevacizumab_11733 raman_Bevacizumab_11733 <NA> 1
4: raman_Bevacizumab_11734 raman_Bevacizumab_11734 <NA> 1
5: raman_Bevacizumab_11735 raman_Bevacizumab_11735 <NA> 1
6: raman_Bevacizumab_11736 raman_Bevacizumab_11736 <NA> 1
7: raman_Bevacizumab_11737 raman_Bevacizumab_11737 <NA> 1
8: raman_Bevacizumab_11738 raman_Bevacizumab_11738 <NA> 1
9: raman_Bevacizumab_11739 raman_Bevacizumab_11739 <NA> 1
10: raman_Bevacizumab_11740 raman_Bevacizumab_11740 <NA> 1
11: raman_Bevacizumab_11741 raman_Bevacizumab_11741 <NA> 1
12: raman_blank_Bevacizumab_10005 raman_blank_Bevacizumab_10005 <NA> 1
13: raman_blank_Bevacizumab_10006 raman_blank_Bevacizumab_10006 <NA> 1
14: raman_blank_Bevacizumab_10007 raman_blank_Bevacizumab_10007 <NA> 1
15: raman_blank_Bevacizumab_10008 raman_blank_Bevacizumab_10008 <NA> 1
16: raman_blank_Bevacizumab_10009 raman_blank_Bevacizumab_10009 <NA> 1
17: raman_blank_Bevacizumab_10010 raman_blank_Bevacizumab_10010 <NA> 1
18: raman_blank_Bevacizumab_10011 raman_blank_Bevacizumab_10011 <NA> 1
19: raman_blank_Bevacizumab_10012 raman_blank_Bevacizumab_10012 <NA> 1
20: raman_blank_Bevacizumab_10013 raman_blank_Bevacizumab_10013 <NA> 1
21: raman_blank_Bevacizumab_10014 raman_blank_Bevacizumab_10014 <NA> 1
22: raman_blank_Bevacizumab_10015 raman_blank_Bevacizumab_10015 <NA> 1
analysis replicate blank spectra
traces
<num>
1: 1024
2: 1024
3: 1024
4: 1024
5: 1024
6: 1024
7: 1024
8: 1024
9: 1024
10: 1024
11: 1024
12: 1024
13: 1024
14: 1024
15: 1024
16: 1024
17: 1024
18: 1024
19: 1024
20: 1024
21: 1024
22: 1024
traces
Results empty
# setting interactive to TRUE, plotly is used for an interactive plot
raman$plot_spectra(interactive = FALSE)
Editing analyses set
For data processing, the analysis replicate names and the correspondent blank analysis replicates can be assigned with dedicated methods, as shown below. For instance, the replicate names are used for averaging the spectra in correspondent analyses and the assigned blanks are used for background subtraction, as shown below in @ref(data-processing).
raman$add_replicate_names(c(rep("Sample", 11), rep("Blank", 11)))
raman$add_blank_names(rep("Blank", 22))
# The replicate names are modified and the blanks are assigned
raman
RamanEngine
name NA
author NA
file NA
date 2024-08-01 10:06:21.050755
Workflow empty
Analyses
analysis replicate blank spectra traces
<char> <char> <char> <num> <num>
1: raman_Bevacizumab_11731 Sample Blank 1 1024
2: raman_Bevacizumab_11732 Sample Blank 1 1024
3: raman_Bevacizumab_11733 Sample Blank 1 1024
4: raman_Bevacizumab_11734 Sample Blank 1 1024
5: raman_Bevacizumab_11735 Sample Blank 1 1024
6: raman_Bevacizumab_11736 Sample Blank 1 1024
7: raman_Bevacizumab_11737 Sample Blank 1 1024
8: raman_Bevacizumab_11738 Sample Blank 1 1024
9: raman_Bevacizumab_11739 Sample Blank 1 1024
10: raman_Bevacizumab_11740 Sample Blank 1 1024
11: raman_Bevacizumab_11741 Sample Blank 1 1024
12: raman_blank_Bevacizumab_10005 Blank Blank 1 1024
13: raman_blank_Bevacizumab_10006 Blank Blank 1 1024
14: raman_blank_Bevacizumab_10007 Blank Blank 1 1024
15: raman_blank_Bevacizumab_10008 Blank Blank 1 1024
16: raman_blank_Bevacizumab_10009 Blank Blank 1 1024
17: raman_blank_Bevacizumab_10010 Blank Blank 1 1024
18: raman_blank_Bevacizumab_10011 Blank Blank 1 1024
19: raman_blank_Bevacizumab_10012 Blank Blank 1 1024
20: raman_blank_Bevacizumab_10013 Blank Blank 1 1024
21: raman_blank_Bevacizumab_10014 Blank Blank 1 1024
22: raman_blank_Bevacizumab_10015 Blank Blank 1 1024
analysis replicate blank spectra traces
Results empty
raman$plot_spectra(interactive = FALSE, colorBy = "replicates")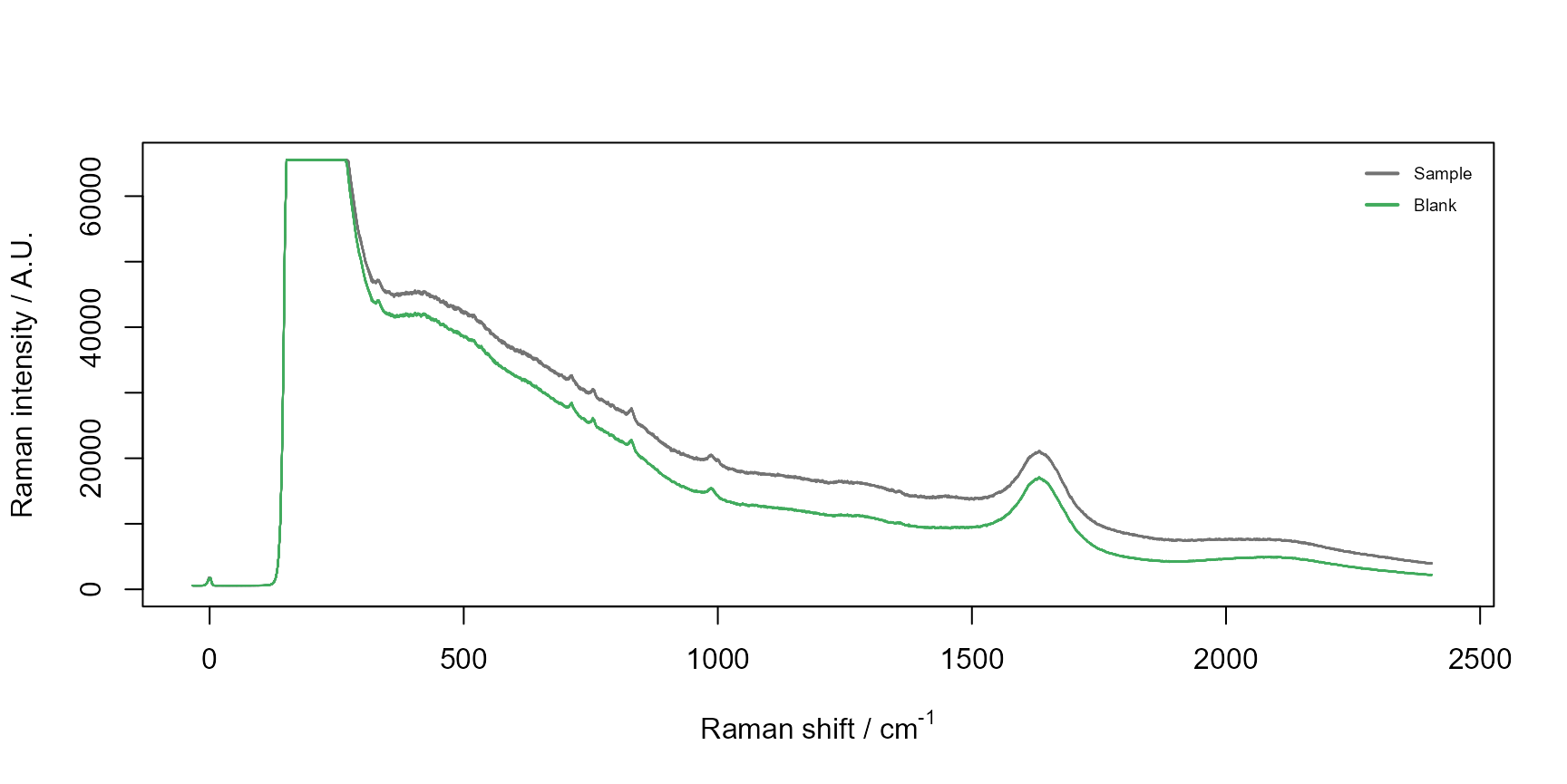
Data processing workflow
As above mentioned, ProcessingSettings are used to
design an ordered data processing workflow. Below we create a simple
list of ProcessingSettings for processing the Raman
spectra.
ps <- list(
# Averages the spectra for each analysis replicate
RamanSettings_AverageSpectra_StreamFind(),
# Simple normalization based on maximum intensity
RamanSettings_NormalizeSpectra_minmax(),
# Background subtraction
RamanSettings_SubtractBlankSpectra_StreamFind(),
# Applies smoothing based on moving average
RamanSettings_SmoothSpectra_movingaverage(windowSize = 4),
# Removes a section from the spectra from -40 to 470
RamanSettings_DeleteSpectraSection_StreamFind(list("shift" = c(-40, 300))),
# Removes a section from the spectra from -40 to 470
RamanSettings_DeleteSpectraSection_StreamFind(list("shift" = c(2000, 3000))),
# Performs baseline correction
RamanSettings_CorrectSpectraBaseline_baseline(method = "als", args = list(lambda = 3, p = 0.06, maxit = 10)),
# Performs again normalization using minimum and maximum
RamanSettings_NormalizeSpectra_minmax()
)
# The settings are added to the engine but not yet applied
raman$add_settings(ps)
raman$print_workflow()
Workflow
1: AverageSpectra (StreamFind)
2: NormalizeSpectra (minmax)
3: SubtractBlankSpectra (StreamFind)
4: SmoothSpectra (movingaverage)
5: DeleteSpectraSection (StreamFind)
6: DeleteSpectraSection (StreamFind)
7: CorrectSpectraBaseline (baseline)
8: NormalizeSpectra (minmax)
# The data processing workflow is applied
raman$run_workflow()Results
Once the data processing methods are applied, the results can be
accessed with the get_results() method.
# The spectra results is added after processing the Raman spectra
raman
RamanEngine
name NA
author NA
file NA
date 2024-08-01 10:06:21.050755
Workflow
1: AverageSpectra (StreamFind)
2: NormalizeSpectra (minmax)
3: SubtractBlankSpectra (StreamFind)
4: SmoothSpectra (movingaverage)
5: DeleteSpectraSection (StreamFind)
6: DeleteSpectraSection (StreamFind)
7: CorrectSpectraBaseline (baseline)
8: NormalizeSpectra (minmax)
Analyses
analysis replicate blank spectra traces
<char> <char> <char> <num> <num>
1: raman_Bevacizumab_11731 Sample Blank 1 1024
2: raman_Bevacizumab_11732 Sample Blank 1 1024
3: raman_Bevacizumab_11733 Sample Blank 1 1024
4: raman_Bevacizumab_11734 Sample Blank 1 1024
5: raman_Bevacizumab_11735 Sample Blank 1 1024
6: raman_Bevacizumab_11736 Sample Blank 1 1024
7: raman_Bevacizumab_11737 Sample Blank 1 1024
8: raman_Bevacizumab_11738 Sample Blank 1 1024
9: raman_Bevacizumab_11739 Sample Blank 1 1024
10: raman_Bevacizumab_11740 Sample Blank 1 1024
11: raman_Bevacizumab_11741 Sample Blank 1 1024
12: raman_blank_Bevacizumab_10005 Blank Blank 1 1024
13: raman_blank_Bevacizumab_10006 Blank Blank 1 1024
14: raman_blank_Bevacizumab_10007 Blank Blank 1 1024
15: raman_blank_Bevacizumab_10008 Blank Blank 1 1024
16: raman_blank_Bevacizumab_10009 Blank Blank 1 1024
17: raman_blank_Bevacizumab_10010 Blank Blank 1 1024
18: raman_blank_Bevacizumab_10011 Blank Blank 1 1024
19: raman_blank_Bevacizumab_10012 Blank Blank 1 1024
20: raman_blank_Bevacizumab_10013 Blank Blank 1 1024
21: raman_blank_Bevacizumab_10014 Blank Blank 1 1024
22: raman_blank_Bevacizumab_10015 Blank Blank 1 1024
analysis replicate blank spectra traces
Results
1: spectra
# The structure of the spectra results
str(raman$get_results("spectra"))List of 1
$ spectra:List of 2
..$ Blank :List of 1
.. ..$ spectra:Classes 'data.table' and 'data.frame': 0 obs. of 0 variables
.. .. ..- attr(*, ".internal.selfref")=<externalptr>
..$ Sample:List of 1
.. ..$ spectra:Classes 'data.table' and 'data.frame': 690 obs. of 6 variables:
.. .. ..$ replicate: chr [1:690] "Sample" "Sample" "Sample" "Sample" ...
.. .. ..$ shift : num [1:690] 300 303 306 309 312 ...
.. .. ..$ intensity: num [1:690] 0.0886 0.0386 0.1075 0.1457 0.2491 ...
.. .. ..$ blank : num [1:690] 0.75 0.733 0.717 0.706 0.695 ...
.. .. ..$ baseline : num [1:690] 0.0454 0.0456 0.0458 0.0461 0.0463 ...
.. .. ..$ raw : num [1:690] 0.0452 0.0453 0.0458 0.0461 0.0467 ...
.. .. ..- attr(*, ".internal.selfref")=<externalptr>
# Averaged spectra can be obtained with the dedicated field
raman$spectra$Blank
Null data.table (0 rows and 0 cols)
$Sample
replicate shift intensity blank baseline raw
<char> <num> <num> <num> <num> <num>
1: Sample 300.2333 0.08860028 0.74995276 0.04535583 0.04520539
2: Sample 303.2319 0.03860228 0.73266994 0.04560113 0.04528911
3: Sample 306.2275 0.10750530 0.71749096 0.04584628 0.04575694
4: Sample 309.2201 0.14567355 0.70615152 0.04609087 0.04612487
5: Sample 312.2137 0.24908625 0.69497025 0.04633435 0.04670256
---
686: Sample 1990.7185 0.13597257 0.06158937 0.04635464 0.04635729
687: Sample 1992.6790 0.13359305 0.06209468 0.04625501 0.04624998
688: Sample 1994.6373 0.12759965 0.06210308 0.04615509 0.04613068
689: Sample 1996.5951 0.11742477 0.06252161 0.04605499 0.04599770
690: Sample 1998.5507 0.11607689 0.06278336 0.04595484 0.04589319
# Modified spectra which results in a single spectrum
raman$plot_spectra()Conclusion
This quick guide introduced the general framework behind StreamFind.
The StreamFind is a data processing workflow designer that uses R6
classes to manage, process, visualize and report data within a project.
The CoreEngine is the parent class of all other data
specific engines and manages the project information via the class
ProjectHeaders. The ProcessingSettings are
used to harmonize the diversity of processing methods and algorithms
available. The data processing is delegated to child engines, such as
the RamanEngine and MassSpecEngine. The data
processing workflow is assembled by combining different processing
methods in a specific order. The results can be accessed with the
get_results() method or dedicated methods
(e.g. spectra and plot_spectra). StreamFind
can also be used via the embedded shiny app for a graphical user
interface. See the StreamFind
App Guide for more information.